OrthoFinder Tutorials Phylogenetic orthology inference for comparative genomics
A2. Getting OrthoFinder input data
Proteomes may be available in a number of places. The (Running an example OrthoFinder analysis) showed how to download data from Ensembl, here are a few pointers for other popular websites.
Phytozome
-
You’ll first need to create an account and log in.
-
Go to the download section:
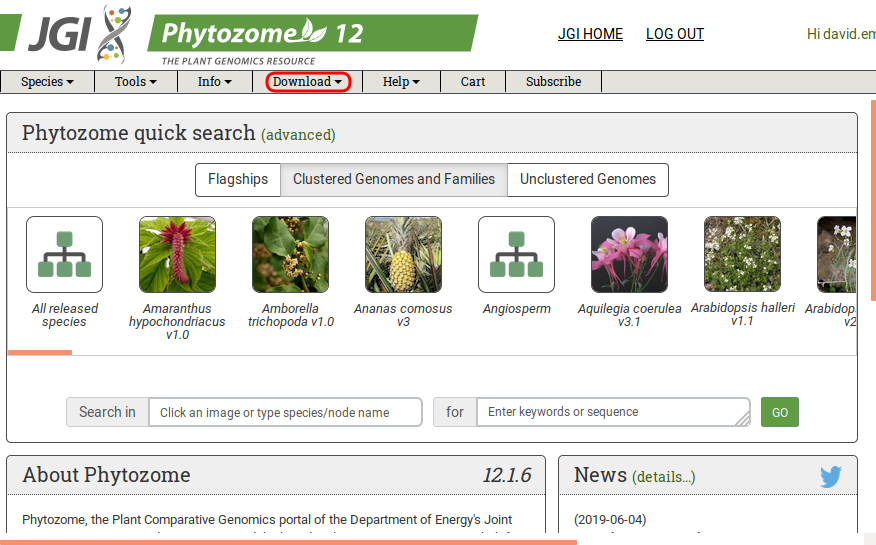
-
Select the _primaryTranscriptOnly.fa.gz files for each species you want.
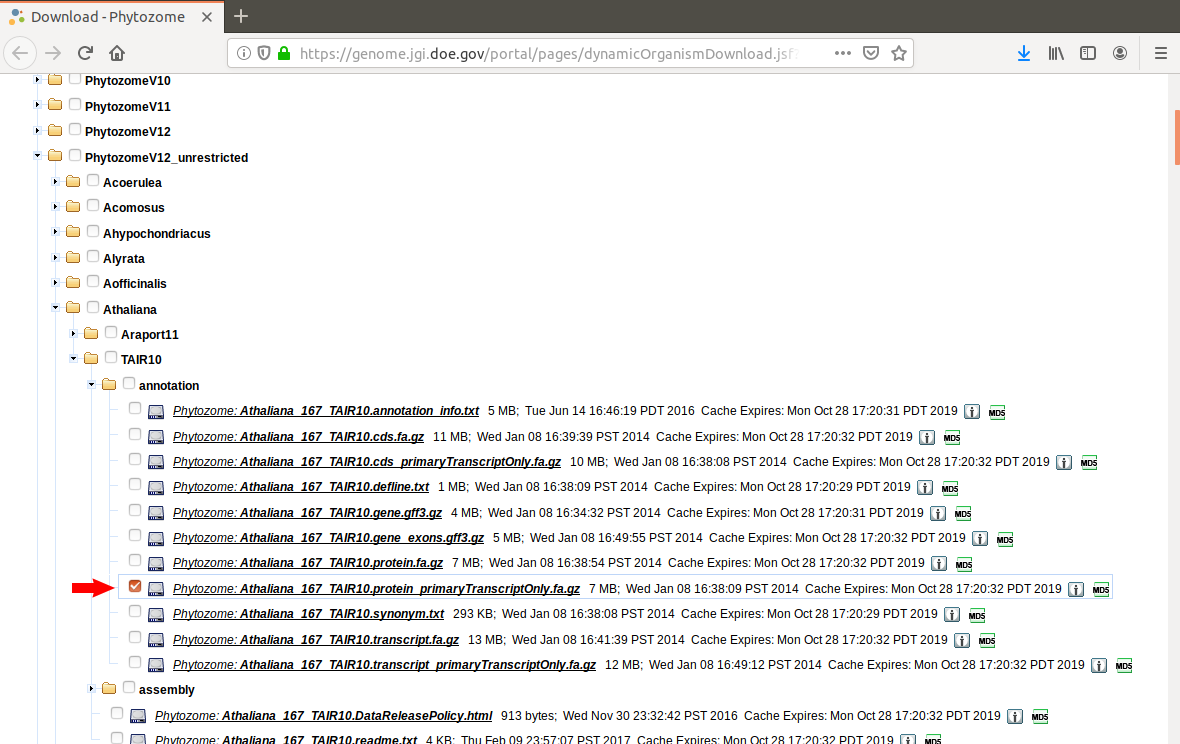
-
Go to the top of the page, and click “Download Selected Files”. They’ll be in a package with the original directory structure so you’ll have to get each of the files, put them in a single file and gunzip them. You don’t need to run the
primary_transcript.pyscript on them as Phytozome has already done that step.
Others?
Let me know if you think pointers to any other sites would be helpful.
Written on October 20th, 2019 by David EmmsFeel free to share!